GONUTS has been updated to MW1.31 Most things seem to be working but be sure to report problems.
evidence codes
|
CACAO the intercollegiate annotation competition!
|
| The Community Assessment of Community Annotation with Ontologies (CACAO) is a competition for teams of undergrads around the world to improve the functional annotation of genes. CACAO was developed and is currently run at Texas A&M University, along with many other institutions. If you are interested in participating, please email us - ecoliwiki@gmail.com. |
This page is to help CACAO students select the correct evidence code & then use that evidence code properly.
Consult the GO consortium evidence guide for the official documentation. This contains the most detailed information on ALL evidence codes, even the ones not permitted in CACAO.
Contents
Evidence Code Overview
- An evidence code is used to describe what type of evidence you are using to support your GO annotation
- We divide them into 5 basic families of evidence codes used by the GO Consortium:
- Experimental
- Computational
Author statementCurator assignedAutomatically assigned (IEA:Inferred from Electronic Annotation)
- CACAO students may only use a subset of Experimental & Computational evidence codes. You do not need to worry about any terms in the crossed-out families.
Experimental
- These are used for capturing experiments performed & published by the authors.
- Your choices for CACAO:
- IDA: Inferred from Direct Assay
- IMP: Inferred from Mutant Phenotype
- IGI: Inferred from Genetic Interaction
- Note: The Inferred in these codes indicates the authours came to some conclusion based on the evidence in the paper, not something the reader infers based on a combination with outside information, etc. Although this rule may have some exceptions, such as gleaning GO:0005624 membrane fraction from a cell fractionation via centrifugation, most of the conclusions you will annotate will be directly stated by the authours.
IDA vs IMP
- IMP trumps IDA. If the authors are using mutants to show a difference between alleles, even if the authors are using a direct assay, use IMP. It is the comparison of the function (or process or component) of the wild-type compared to the mutant.
- If a gene/protein is put in another organism but is still being analysed as if it is in the native organism (no comparison of alleles), is it likely IDA. Remember, a human protein injected into a mouse is still a human protein.
IMP and regulation
Changes in the level or activity of protein Y when protein X is mutated can be used to infer that protein X regulates some aspect of protein Y. Note, however:
- Regulation of expression is not a subset of regulation of activity. Use "regulation of X activity" terms when the mutation affects the specific activity of the target. For example, regulatory subunits of enzymes should be annotated with regulates->activity terms. Transcription factors that control expression of enzymes should not.
- Changes in mRNA levels do not always mean that regulation of gene expression annotations can be made. Beware of indirect effects. Note also that GO annotation is based on capturing what is in the literature, not what you as a curator infer from the data. Your inference did not pass peer review! So if the authors report that the mRNA levels of several genes changes in a microarray experiment where gene X is mutated, only annotate that that gene X regulates the processes associated with the regulated genes if there are also definitive author statements that made it through peer review.
- For example: "this shows that X regulates..." is definitive.
- "this suggests that X could regulate..." is not definitive.
- For example: "this shows that X regulates..." is definitive.
- If you want to annotate X as regulating a biological process based on its effects on expression of genes that are involved in that process, first make sure those genes are experimentally annotated to the process.
IGI
- This interaction term needs the with/from field filled in, but you get two (or more!) annotations for the price of one.
- You can use IGI if a mutation in one gene makes the authours draw a conclusion about another gene.
- It is more common to use this if the authors have to make multiple mutations in the same experiment to see a change in phenotype (i.e. there are 2 genes that code for function X, so you have to mutate both to eliminate function X in the organism).
- What goes in the with/from field on the protein page for Gene X is the UniProt (UniProtKB) accession for the OTHER gene (gene Y) the authors mutated.
Note: If the accession in the With/From matches the accession of the protein page, the annotation is WRONG.
The best part is that you can make the exact same annotation on the OTHER (Y) protein's page, but use the UniProt (UniProtKB) accession from your first protein (gene X) in the with/from!
This paper is about how the authors needed to knockout both fabA & fadD genes in E. coli to disrupt its fatty acid biosynthesis.
On the fabA protein page from E. coli page, this is what the final annotation looks like:
Computational
- These are capturing amino acid sequence studies published by the authors
- You'll probably seldom use these, and other than ISA, they are considerably tricker and harder to understand than the Experimental codes.
Your choices for CACAO:
- ISA: Inferred from Sequence Alignment
- ISO: Inferred from Sequence Orthology
- ISM: Inferred from Sequence Model
- IGC: Inferred from Genomic Context
ISA
Example-Using ISA in a GO annotation:
- The authors publish an amino acid sequence alignment of a protein of interest to 1 or more other proteins & say your protein aligns well with these other proteins, which are known to be cysteine proteases. You would like to make an annotation for your protein to cysteine protease activity (or similar GO term) because of this, but you will have to use ISA correctly.
- You must fill in the with/from with the UniProt (UniProtKB) accession for one of the other proteins (this is not the hard part)
- BUT this other protein(s) must have an experiment-coded (IDA, IMP, IGI, IEP, etc... NOT IEA) annotation to cysteine protease activity (this IS the hard part)
- Once you identify the Uniprot accession for the other proteins, use the {Special:Create_New_Gene_Page "Create New Gene Page" maker] on GONUTS. If there is no existing page, this will create one, and it is likely you will not be able to use that protein as a with/from "reference". If there is already a page, the Page Maker will direct you to it.
- Examine ALL entries on the gene page. If you see one that has the EXACT term you intend to annotate to, and has an acceptable evidence code (again, NOT IEA), then you can use
Here is an actual example taken from Chua et al. (1988), where the authors are aligning DerP1 from the European Dust Mite to other known cysteine proteases:
If you want to make an ISA annotation on the DerP1 page, you have to find out if any of these other proteins have an annotation to the EXACT TERM based on an experiment. Happy hunting - here's step by step instructions:
Step 1. Make sure your paper has a sequence alignment where the authours explicitly state there is a strong alignment
Step 2: Go to UniProt & look up the accessions for each of these other proteins the authours state your gene is similar to
rat cathepsin H = P00786
no accession for Chinese Gooseberry actinidin
Papaya papain = P00784
human cathepsin B = P07858
Step 3: Click on each accession & scroll down to the section called "Ontologies"
Step 4: Click on the link that says "Complete GO annotation..." for each protein.
(FYI, this is what GONUTS gets from UniProt when you make a page for a protein on GONUTS ).
Step 5: Examine ALL entries on the gene page.
* If you see one that has the '''EXACT''' term you intend to annotate to, and has an acceptable evidence code (again, ''NOT IEA''),
then you can use that ONE Uniprot accession number in the with/from field.
* If more than one of the aligned genes have acceptable (remember, '''EXACT''' and '''NOT IEA''') annotations, you can use those too.
Put all the acceptable, aligned genes in the same annotation.
So
* Rat has no molecular function terms that are annotated with experimental evidence codes except an IPI annotation to a binding term,
which CACAO students cannot use. NO to using the rat protein
* Chinese Gooseberry - no accession, thus no GO annotations. NO to using the Chinese Gooseberry protein.
* Papaya has no annotations except to IEA. NO to using papaya protein.
* Human Cathepsin B has an annotation to GO:008234 cysteine-type peptidase activity with IDA as the evidence code
& the reference PMID:7890620.
WE CAN USE THIS ONE!!! THE UNIPROT ACCESSION FOR THIS PROTEIN (P07858) WILL GO IN THE WITH/FROM FOR OUR ANNOTATION ON THE DerP1 PAGE.
- This is what a complete GO annotation looks like using the ISA evidence code. This is on the DerP1 protein page.
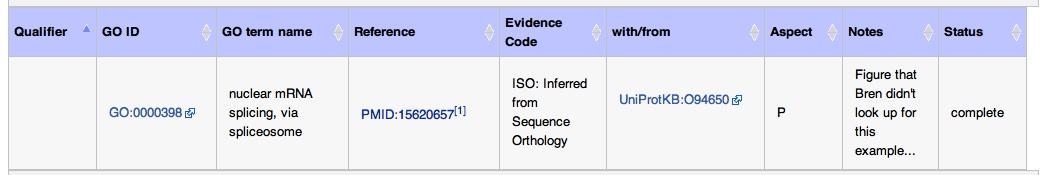
ISO
- This evidence code is similar to ISA, which is why if you are in doubt, use ISA. This evidence code rarely gets used.
- ISO requires the with/from to be filled in with an "other" protein that is being aligned via amino acid sequence.
- Use ISO when your protein and the "other" protein are known to be functional orthologs of each other. This usually involves phylogenitic analysis (maximum likelihood, nearest neighbour joining,...).
- Remember, the referenced protein that is used in the with/from field must have the EXACT term annotated with an evidence code other than IEA
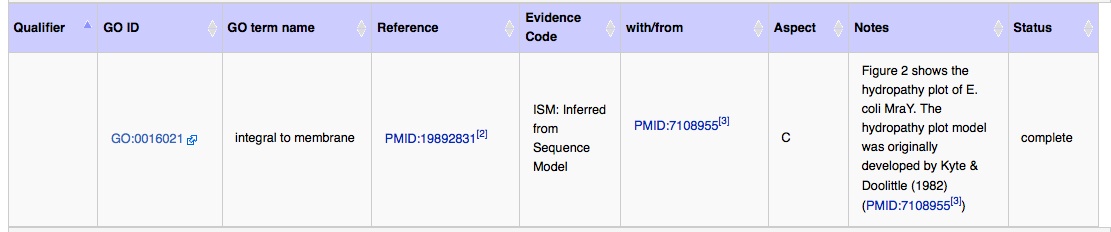
ISM
- This is a funky evidence code because you have to put a PMID in the with/from field.
- This PMID identifies the paper that describes the method used to predict the protein's function.
- You use it when the authors have used a computational modeling program to predict something about your protein (i.e. transmembrane domains via a modeling program called hydropathy plotting, which would mean that the protein is going to be located in the membrane).
Here is an example of correct usage - the annotation is on the E. coli MraY protein page
IGC
The GO consortium describes four situations where genomic context can be used to infer annotations
- operon structure
- syntenic regions
- pathway analysis
- genome scale analysis of processes
These require use of the with/from field, but note that the usage is different depending on the type of IGC evidence. For operons, pathways, or genome scale process analysis, the idea is that you can infer the function of the gene from which other genes are coexpressed or coinherited through evolution.
Thus, use one or more identifiers to specify the other genes in the operon, pathway, or process.
Useful Handouts
If you have not yet visited the helpful handouts for students page, these links may be helpful:
- Evidence code decision tree: A chart with yes/no questions that can be very useful in determining what evidence code to use.
- Experiments and their evidence codes: Has a list of some types of experiments and the evidence codes that are usually associated with them. This is good as a starting point if you're not quite sure what the experiment is showing, but remember, one type of experiment may support different evidence codes; for example what shows an IMP for a process term could possibly be an IDA for a component term as well.
References
See Help:References for how to manage references in GONUTS.