GONUTS has been updated to MW1.31 Most things seem to be working but be sure to report problems.
Help:Annotation Jamborees
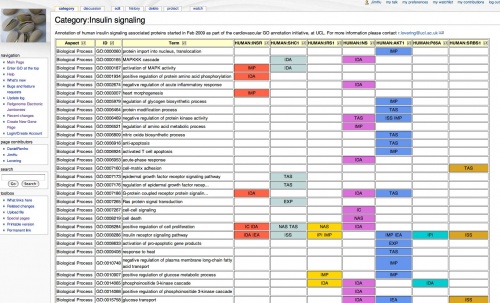
Annotation Jamboree pages are used to compare the annotations of sets of genes of interest. GONUTS allows you to create Category pages that can automate the comparisons. This page describes how to set up a Jamboree page in GONUTS, and gives advice for using it.
Contents
Quick overview
- Create a jamboree category page, e.g. Category:xx
- Create gene pages using the 'create new gene page' on the left hand menu, add the uniprot ID (or refseq protein ID)
- Wait for the site to be created
- The annotations are downloaded automatically
- Newly created annotations need to be added manually to this gene page
- Use the 'edit table' link at the bottom of the annotation table
- Add row (using button at bottom of editing annotation table)
- Save row edits
- Save table when you have finished editing
- Add [[Category:xx]] to the 'notes' section of the gene page, using 'edit' for that section.
Setup
Create the Jamboree Category page
You can use the inputbox below (for RefGenome jamborees the one on Category:Refgenome_Electronic_Jamborees preformats the PAGENAME)
Alternatively, if you don't like that format, you can add the <GOsummary />tag to any category page.
Add genes to the Category
Find the genes you want to include in the Jamboree. You can create gene pages as needed from UniProtKB ids using Special:GoPageMaker.
Add a live chat (optional)
For RefGenome Jamborees, we typically embed a live chat using CoverItLive
<iframe_embed> coveritlive altcast code < iframe_embed >
See Help:Embedding iframes for more info.
Annotate the individual gene pages
Use the table editor to add annotations to each gene page in the category. The jamboree page should automatically update (you may need to reload your browser) to reflect changes in the annotations assigned to each gene.
View the summary on the jamboree category page
An extension finds all the gene pages assigned to this category, filters out the IEAs and builds a comparison table. It also generates images to show how the annotations are distributed among the ontologies.
If you would like to bulk load your annotation data, the format of the input file is:
Qualifier||GOID||GO term name||reference (e.g.PubMed ids) ||evidence code||with data||aspect||notes|| ||
e.g. ||GO:0006919||activation of caspase activity||18082289 ||IDA|| ||P|| || || ||GO:0015684||ferrous iron transport||18082289,17109629,17293870,12734107 ||IDA|| ||P|| || ||
Examples
- Refgenome jamborees. Before July 2009, a different setup was used. The earlier Jamboree pages are individual articles. Starting in July 2009, the Jamboree pages are subcategories.
- Category:Complement_comparison
- Category:Insulin_signaling